Mom and iPOP
At the age of 54, at Stanford University had a project that required his mother’s help. The project, which would catapult Snyder into the headlines of science media outlets, was the , iPOP. Snyder and his team were gathering genomic, transcriptomic, proteomic, metabolomic and autoantibody analyses from one person to see what kinds of information could be obtained by integrating the statuses of thousands of molecules at once. The project was the first of its kind, aiming to demonstrate that -omics technologies put together can reveal details about a person that a single type of technology can’t accomplish.
The person being sampled was Snyder himself. And because of the choice of sample, Snyder knew his mother’s genetic information could be useful. “She’s a curious person by nature,” says Snyder, so he described to her over the phone what he was attempting to do.
“Every mother likes to help out their child,” says 84-year-old Phyllis Snyder. “I figured if Mike needs help, I’d be happy to help him. Plus, I was a chemistry major first before I became a teacher. I always think, ‘anything for science!’”

The me-ome
The iPOP project had been mulling around in Mike Snyder’s head since 2004, when his group was based at Yale University and working on genomics, transcriptomics and proteomics as individual areas of research. Snyder, like other molecular biologists, was keenly aware that genes, RNA transcripts, proteins and other molecules don’t operate in isolation; much can be learned from the intricate connections between these different classes of molecules. Inevitably, the thought started to crop up in Snyder’s head — what if all the different types of -omics could be analyzed at once?
In 2009, technologies and advances in molecular biology matured to a point that it was time to take a stab at the idea, says Snyder. The technology to sequence a person’s genome already was in place, and Snyder’s group had the expertise to tackle the transcriptomics, proteomics and metabolomics. Snyder got his whole genome sequenced through several different approaches. Over 14 months, Snyder had his transcriptomic, proteomic, metabolomic and autoantibody profiles collected.
His mother’s genome was going to be useful to determine what Snyder had or hadn’t inherited from her. Snyder asked his mother if she was willing to have her genome sequenced and explained the consequences and ethical considerations of genome sequencing. “I said, ‘Sure! Anything you need!’” Phyllis Snyder recalls. “Then he said he would like some blood.”
Her son sent her the consent forms and enrolled her in the study. Phyllis Snyder visited her doctor to have him draw her blood sample and take a buccal swab. She sent the materials to her son. “And that was it!” she says. Asked if she was worried about what her genome might reveal, Snyder says, “I am 84 years old. If something hadn’t shown up before and it turns up now, well, what can you say? I’ve had a good life and I am very proud of all my children.”
And so the mother-son duo appeared in a paper in Cell (1). The paper described details captured by Mike Snyder’s iPOP that couldn’t be seen with standard clinical diagnostics. For example, during those 14 months, Snyder began to develop the symptoms of diabetes, high blood sugar and a particular form of glycosylated hemoglobin. Snyder didn’t have any of the physical predispositions for diabetes: He wasn’t overweight, didn’t smoke and didn’t have a family history of the illness. But as his iPOP began to display the telltale signs and he received the diagnosis for the disease, Snyder immediately took action to cut off the disease’s progress. “No more junk food or desserts,” he says. “Cake, ice cream, candy all gone.” He doubled the amount of biking he does and started running again. With these changes, his iPOP later showed a drop in the diabetic biomarkers.
iPOP also demonstrated how one data set may not be sufficient for the whole picture. The genomic analyses revealed that both Snyders had a specific mutation that has been correlated with aplastic anemia, a condition in which the body doesn’t produce enough new red blood cells. But neither of them has the disease, indicating that genetics alone isn’t enough to kick off this particular disease.
The iPOP experiment now has Mike Snyder interested in studying the -omics profiles of prediabetics and seeing what information can be gathered if they cross over into the realm of diabetes. His group now is working to get the requisite approvals in place to carry out iPOP analyses on 250 prediabetic patients. “One-third will convert to diabetics over the course of five years,” says Snyder. “We hope to learn what triggers this conversion and what the final disease profile looks like. We also want to learn why some people respond to some drugs and others do not.” He speculates that diabetes could be many diseases rolled into one; through iPOP, he hopes his team will be able to discern exactly how many metabolic pathways go awry and how people might be best treated.
Snyder aims to make iPOP a more readily accessible tool for health-care providers. He envisions a future when everyone can get an iPOP done for $100 or so, “the same cost as the blood test you do now,” he says. The costs associated with an iPOP should come down over time, because researchers should become more adept at knowing which molecules should be tracked and which ones are not worth following, says Snyder. By picking out the most informative biomarkers, an iPOP should become more streamlined and cost effective.
 Michael Snyder with his youngest child, Eve.
Michael Snyder with his youngest child, Eve.
Catching the science bug
Phyllis Snyder was the first person to get her son interested science. She was a stay-at-home mother of six children and went through college part time when Snyder was young and joined the work force to help support the family. She began to work as a substitute elementary school teacher when Snyder was in high school and became a full-time third-grade teacher when Snyder entered college. He credits her for bestowing an inquisitive mind in him and his siblings as well as a sunny outlook on life. “She is a positive person by nature,” says Snyder, “and I like to think I am.”
Snyder’s father passed away in 2000. An accountant, Kermit Snyder was “extremely good with numbers and a great dad,” says Snyder, adding that he and his siblings “got some of our quantitative aspects from him.” Two of Mike Snyder’s siblings also have Ph.D.s., one in organic chemistry and the other in psychology. As Phyllis Snyder quips, “Three doctors, but none of them can cure you of anything!”
Snyder is the fifth of three boys and three girls and reckons family dynamics taught him a thing or two about collaboration and competition. “In a large family, you learn how to get along with others as well as stand up for yourself,” he says. “I remember at the dinner table that if you hesitated you would not get dessert because it would all be eaten.”
His mother describes Snyder as “energetic and a workaholic, like his father” as well as “very considerate and compassionate for others.” When Snyder was in elementary school, Phyllis Snyder says, the family called him “the absentminded professor.” “We would all go crazy looking for his shoes before he went to school, because he couldn’t remember where he had left them,” she recalls, laughing.
His childhood home was modest, says Mike Snyder. His family lived just outside a town called Pottstown in Pennsylvania dairy land in a farmhouse that was more than 100 years old with sprawling grounds. “Most people are born, raised and die within a 15-mile radius,” he says. Local interests are geared more toward sports than academics.
A high-school chemistry teacher, Mr. Darby (“I don’t know his first name,” says Snyder), also fed Mike Snyder’s interest in the scientific enterprise. Snyder remembers the freedom he was given in Darby’s advanced chemistry class to explore science through independent research projects. Snyder went on to win a Bausch & Lomb science award, which included a free application to the University of Rochester. Although Snyder applied to other schools, the University of Rochester offered him a scholarship. “We didn’t have a lot of money,” says Snyder, “so I went there.”
 Phyllis Snyder with her granddaughter Eve, the youngest of 11 grandchildren.
Phyllis Snyder with her granddaughter Eve, the youngest of 11 grandchildren.
Snyder picked chemistry as his undergraduate major, but toward the end of college he developed an interest in biology. Although Snyder got accepted for graduate school in chemistry, “I decided to take a year off to learn biology and work in a biology research laboratory,” he says. Soon he was doing his graduate work from 1978 to 1982 with molecular biologist at the California Institute of Technology and his postdoctoral training for four years with at Stanford University. Davis is now a colleague at Stanford and also trained under Davidson.
Snyder says Davidson and Davis served as mentors on how get science done. Davidson taught Snyder how to think independently and analytically. Davis showed Snyder the power of thinking outside the box. “Whatever I’ve learned from Norman and Ron, it’s been valuable,” notes Snyder.
Davis recalls Snyder being a hard-core carnivore in his younger days. “He used to say, ‘I hate vegetables.’ I was a vegetarian at that time so I said, ‘We complement one another very well!’”
The complementarity extended beyond food. “I admire him a great deal,” says Davis. “We get along well partly because of our similar mindset.” The mindset, Davis says, is a legacy from Davidson, who firmly believed biology would always advance with novel and improved technologies.
Davis says that Snyder’s approach to science hasn’t changed since his postdoctoral days with him. Instead of copying what other researchers were pursuing, Davis says, Snyder always identified the next big question that wasn’t being explored and went after it. “That attitude represents his whole career path,” notes Davis.
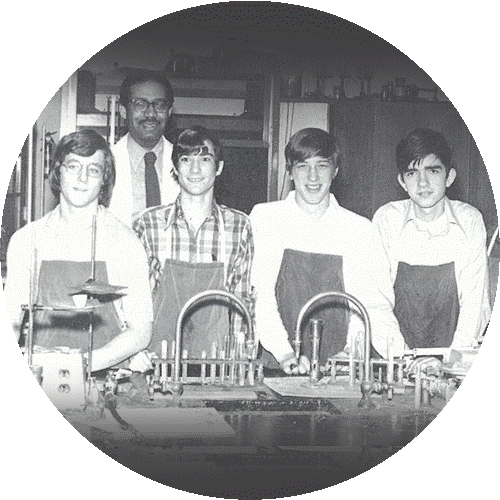 Michael Snyder (second student from left) credits his advanced chemistry teacher, Mr. Darby (back row), with giving him the freedom to explore science through independent research projects.
Michael Snyder (second student from left) credits his advanced chemistry teacher, Mr. Darby (back row), with giving him the freedom to explore science through independent research projects.
Bridging chemistry and biology
The iPOP work showcases Snyder’s longstanding interest in understanding what differs between people at the molecular level. “We’re all really chemical reactions. Understanding the basic laws of chemistry will go a long way toward defining what we are,” he says. He feels the mission of a molecular biologist is to understand how chemical reactions in our bodies differ in their responses to genetic and environmental stimuli. He hopes that researchers soon will become so good at interpreting molecular details that they can use those details to make predictions of how a person with a particular genomic variant will respond to a certain environmental event.
Before iPOP came along, Snyder’s group at Yale University spent considerable effort learning about variation at the transcription level. Snyder cites one of the key findings his group made in yeast when they demonstrated that, contrary to popular belief, the regulatory sequences were not conserved. “There were extensive differences in transcription factor binding even amongst closely related species,” says Snyder. “It took three years to get that paper published, because most people didn’t believe it.” (2) Snyder says that growing evidence “really hammers home the argument that we differ more at the level of regulation than simply coding sequences.”
Snyder’s group also has made inroads into functional genomics and proteomics. They demonstrated the power of using microarrays of transcripts and peptides to study gene expression patterns and protein phosphorylation. Snyder’s laboratory was the first to perform a large-scale functional genomics project in any organism and also created the first proteome chip. They carried out the first high-resolution tiling array for the entire human genome and also spearheaded the use of high-throughput DNA sequencing technologies to study bacterial genomes. Snyder’s laboratory at Stanford, to which he moved three years ago, continues to focus on a variety of projects in the areas of genomics and proteomics both in yeast and humans.
Snyder has donned other hats other than that of a scientist. Between 1998 and 2004, he was chairman of the department of molecular, cellular and developmental biology at Yale, during which time the department doubled in size and tripled its funds. He is the chairman of the genomics center at Stanford now, and Davis credits him with setting up a sophisticated sequencing center and working to introduce genome sequencing into clinical practice by partnering with the Stanford University Medical Center. Snyder is also an associate editor for (a publication of the 91—«…´¥´√Ω). He has experience on the biotechnology side of science through service on various scientific advisory boards and as a founder of five companies.
Snyder’s enthusiasm for science is unrestrained. “Every morning, I tell my kids, ‘I’m going to fun!’ They always tease me back and say, ‘No, you’re not. You’re going to work!’” says Snyder. But he truly means it when he calls work fun. “That’s why I do science. I love it.”
As in every parent-child relationship, Phyllis Snyder gets the last word. “Every mother thinks her son is brilliant,” she says. “And I am no different.”
References
- Chen, R., et al. Cell148, (2012).
- Borneman, A.R., et al. Science317, (2007).
See a presentation by Snyder on “Genomics and Personalized Medicine,” presented in October 2009 and made available as part of Stanford University’s .
Editor's note (added July 19, 2012)
A keen reader pointed out that Phyllis Snyder is listed on the Cell paper as an author with an affiliation to Stanford University, so we followed up with Michael Snyder to find out why. He says that he and his co-authors felt that Phyllis Snyder deserved authorship because she provided information and samples that were critical to the study. He adds that Phyllis Snyder’s affiliation with Stanford was a mistake that he hadn’t caught when the paper was submitted to the journal.
Enjoy reading 91—«…´¥´√Ω Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from 91—«…´¥´√Ω Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Unraveling oncogenesis: What makes cancer tick?
Learn about the 91—«…´¥´√Ω 2025 symposium on oncogenic hubs: chromatin regulatory and transcriptional complexes in cancer.

Exploring lipid metabolism: A journey through time and innovation
Recent lipid metabolism research has unveiled critical insights into lipid‚Äìprotein interactions, offering potential therapeutic targets for metabolic and neurodegenerative diseases. Check out the latest in lipid science at the 91—«…´¥´√Ω annual meeting.

Melissa Moore to speak at 91—«…´¥´√Ω 2025
Richard Silverman and Melissa Moore are the featured speakers at the 91—«…´¥´√Ω annual meeting to be held April 12-15 in Chicago.

A new kind of stem cell is revolutionizing regenerative medicine
Induced pluripotent stem cells are paving the way for personalized treatments to diabetes, vision loss and more. However, scientists still face hurdles such as strict regulations, scalability, cell longevity and immune rejection.

Engineering the future with synthetic biology
Learn about the 91—«…´¥´√Ω 2025 symposium on synthetic biology, featuring applications to better human and environmental health.

Scientists find bacterial ‘Achilles’ heel’ to combat antibiotic resistance
Alejandro Vila, an 91—«…´¥´√Ω Breakthroughs speaker, discussed his work on metallo-Œ≤-lactamase enzymes and their dependence on zinc.

