Our evolving view of plasma membrane domains
We recently used a new chemical imaging technique to visualize the and in the plasma membranes of fibroblasts. Our unexpected finding of cytoskeleton-dependent sphingolipid domains that are not enriched with cholesterol has led us to revise our views on plasma membrane domains.
Though the plasma membrane may contain microdomains with a variety of different lipid compositions, cholesterol- and sphingolipid-enriched microdomains, called lipid rafts, have been the most intensely scrutinized. Lipid rafts are defined as small, dynamic and ordered assemblies of cholesterol, sphingolipids and proteins . Rafts are postulated to regulate protein–protein interactions by laterally segregating proteins according to their affinity for ordered membrane domains. Cell signaling and virus budding are among the processes that lipid rafts hypothetically mediate. This potential importance and the simplicity of the raft hypothesis attracted us to the field.
Though membrane organization and function is now our focus, we entered this field as a bioanalytical laboratory that was developing new methods to obtain chemical information from biointerfaces with submicron lateral resolution. At the time, the raft hypothesis was supported strongly by indirect data, including the spontaneous formation of ordered cholesterol- and sphingolipid-enriched domains in model membranes and the cholesterol-sensitive clustering of membrane proteins thought to reside in rafts. Yet domains enriched with cholesterol and sphingolipids had not been imaged directly in actual cell membranes, Techniques for imaging dynamic lipid domains that were smaller than the diffraction limit of light were not yet widespread. Furthermore, cooperative interactions between cholesterol and sphingolipids might be perturbed by labeling them with fluorophores.
To address this challenge, we were developing an approach that used a new surface-sensitive imaging mass-spectrometry technique, high-resolution secondary ion mass spectrometry, or SIMS, to visualize metabolically incorporated, stable isotope-labeled sphingolipids and cholesterol in the plasma membrane with ~100-nm lateral resolution. High-resolution SIMS is complementary to fluorescence microscopy, because stable isotopes do not alter the chemical structure and thus the interactions or trafficking of the lipids they label, but it cannot be performed on living cells.
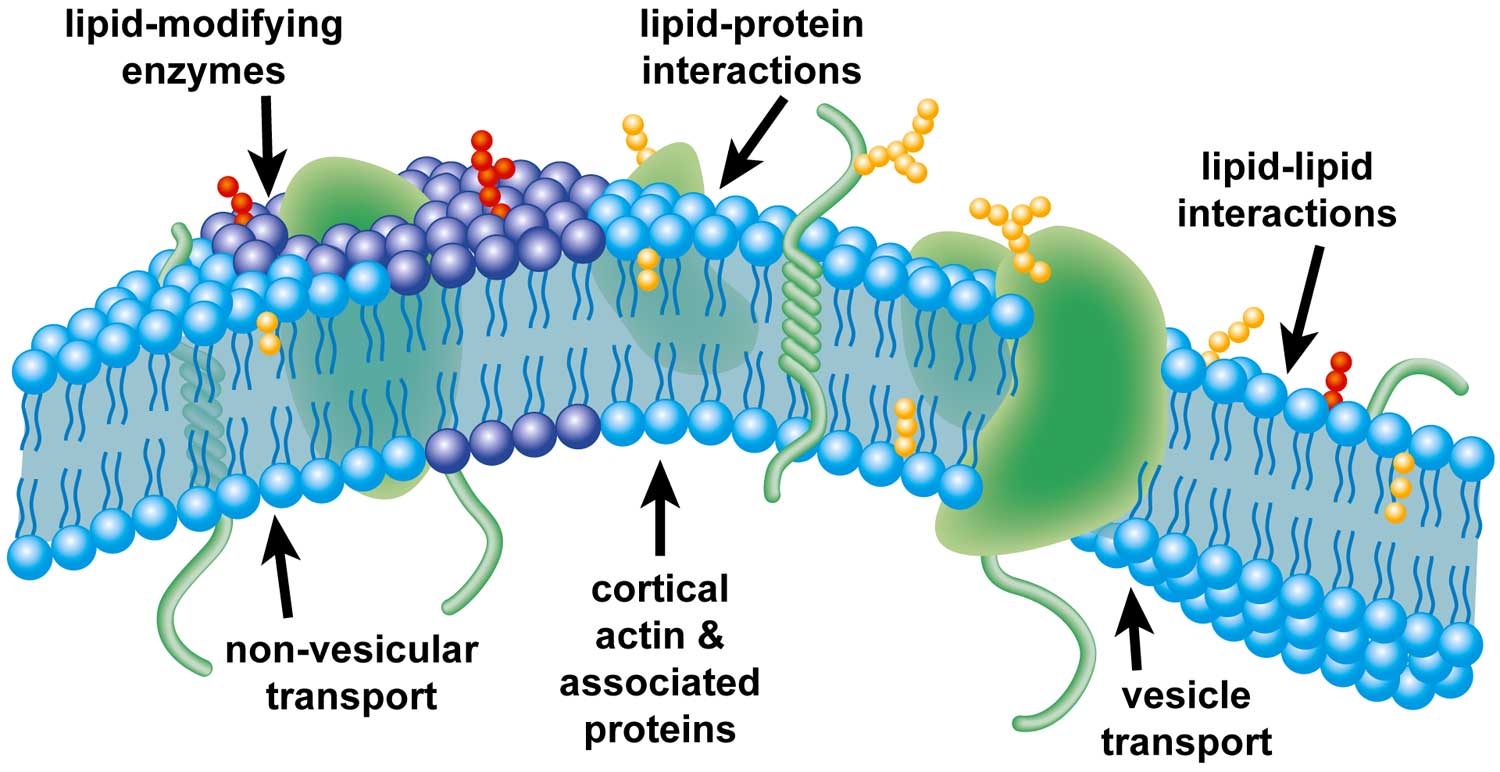 |
| Illustration of factors that may affect the formation of plasma-membrane domains. |
We obtained our first images of the sphingolipid and cholesterol distribution in the plasma membranes of fibroblast cells in 2009. To our surprise, we saw sphingolipid domains that were too large to be lipid rafts and a relatively uniform cholesterol distribution within the plasma membranes of mouse fibroblast cells. Baffled by these results, we spent the next few years optimizing our approach for imaging sphingolipid distribution. After confirming reproducibility, we performed numerous control experiments that tested for a multitude of potential artifacts.
that the micrometer-scale sphingolipid domains we observed were induced by cell fixation, the detection of excess lipid material due to vesicles or intracellular membranes adjacent to the plasma membrane, cell topography, nonspecifically adsorbed labels and many other artifacts. the existence of micron-scale sphingolipid domains in the plasma membrane by fluorescence microscopy imaging of metabolically generated fluorescent sphingolipids in the membranes of living fibroblast cells.
How could our finding of micron-scale sphingolipid domains be correct when it seemed to contradict so much previously reported data? Upon reevaluating the literature, we found that the contradiction was not with previously reported data but instead with the conclusions that had been inferred from the biophysical properties of putative raft components. The few reports in which sphingolipids were imaged directly showed that but that sphingomyelin with dimensions similar to those we observed.
We next probed the mechanisms responsible for these sphingolipid domains. We found that the abundances of the sphingolipid domains in the plasma membrane were reduced by depletion of cellular cholesterol. However, depolymerization of the actin cytoskeleton abolished the sphingolipid domains. The sizes of the sphingolipid domains and their higher dependency on the cytoskeleton than on cholesterol indicated these domains were not lipid rafts.
After publishing , we returned to imaging cholesterol in parallel with sphingolipids. Again, we found that cholesterol was distributed fairly uniformly in the plasma membrane and . This lack of cholesterol enrichment confirmed that the sphingolipid domains were not lipid rafts. Of course, we cannot rule out the possibility that we did not detect lipid rafts because they are smaller than the 87-nm-lateral resolution we achieved. However, the lack of cholesterol enrichment in the sphingolipid domains indicates cohesive cholesterol–sphingolipid interactions contribute little to plasma membrane organization, which refutes a major tenant of the raft hypothesis.
argue against the existence of lipid rafts within the plasma membrane. Instead, our results support the model of plasma membrane organization in which cortical actin and its associated proteins divide the plasma membrane into distinct domains by establishing diffusion barriers that sustain concentration gradients .
Furthermore, the has shown that these gangliosides form separate microdomains that are dependent on the actin cytoskeleton. Thus, microdomains consisting of different sphingolipid species are present in the plasma membrane, which suggests an active, energy-dependent mechanism of membrane organization.
Given that (i.e., ceramide, diacylglycerol, lysophosphatidic acid), cells likely use active mechanisms to segregate each bioactive lipid class within the plasma membrane so that signaling molecules are available when needed. Of course, further studies are required to assess the hypothetical existence of domains of different bioactive lipids and to identify the mechanisms for their formation.
Why do cholesterol levels affect cell function in the absence of cholesterol-enriched plasma-membrane domains? Emerging hypothetical mechanisms of cholesterol-mediated cell function involve modulation of protein activity by direct cholesterol binding. Cholesterol binding/unbinding to scaffold protein to regulate the formation of a signaling complex and its function.
The cholesterol-sensitive assembly of these signaling complexes could be modulated by the difference in the cholesterol concentrations in the plasma membrane and the intracellular locations where the scaffold protein resides. For instance, its co-localization with the cystic fibrosis transmembrane conductance regulator, known as CFTR, at the plasma membrane and NHERF1-mediated CFTR activation.
Likewise, oxysterol-binding protein functions as a scaffold protein that requires cholesterol binding in order to complex with two phosphatases, PP2A and HePTP, . Cleary, much research is required to assess the roles of specific cholesterol–protein interactions in cholesterol-sensitive cell function.
Identifying the mechanisms that control lipid organization within the plasma membrane and the sources of cholesterol-sensitive cellular processes will require a significant number of new studies. We expect that new efforts to develop and test alternative hypotheses for lipid-mediated biological function are critical to advancing our understanding of plasma membrane domains and their roles in cellular function.
Enjoy reading 91СЗЙ«ґ«ГЅ Today?
Become a member to receive the print edition four times a year and the digital edition weekly.
Learn moreGet the latest from 91СЗЙ«ґ«ГЅ Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Meet Robert Helsley
The Journal of Lipid Research junior associate editor studies chronic liver disease and was the first in his family to attend college.

From the Journals: MCP
Protein acetylation helps plants adapt to light. Mapping protein locations in 3D tissues. Demystifying the glycan–protein interactome. Read about these recent papers.

Exploring life’s blueprint: Gene expression in development and evolution
Meet Julia Zeitlinger and David Arnosti — two co-chairs of the 91СЗЙ«ґ«ГЅвЂ™s 2025 meeting on gene expression, to be held June 26-29, in Kansas City, Missouri.

From the journals: JLR
Protein analysis of dopaminergic neurons. Predicting immunotherapy responses in lung cancer. ZASP: An efficient proteomics sample prep method. Read about papers on these topics recently published in Molecular & Cellular Proteomics.

New mass spectrometry assay speeds up UTI diagnosis
Scientists in Quebec use liquid chromatography–mass spectrometry to reduce the time needed to test for bacteria in urine from days to minutes — and with smaller samples.

From the journals: MCP
Protein analysis of dopaminergic neurons. Predicting immunotherapy responses in lung cancer. ZASP: An efficient proteomics sample prep method. Read about papers on these topics recently published in Molecular & Cellular Proteomics.


