
The antibody patent question
The U.S. Supreme Court will decide in early 2019 whether to consider a patent dispute over monoclonal antibodies. At issue is whether a company, namely the pharmaceutical giant Amgen, can patent every antibody that binds a given epitope — even if there might be thousands of such antibodies and not all are yet discovered.
Amgen filed the patents in question in 2008. They cover a class of antibodies that bind to the proprotein convertase subtilisin/kexin type 9, or PCSK9, and thereby lower cholesterol. Amgen markets one PCSK9 inhibitor as Repatha. The company is suing rival pharmaceutical giant Sanofi over its competing drug, Praluent, which targets the same part of PCSK9.
If Amgen goes before the high court and prevails, it will get to lay claim to a whole therapeutic approach. If it loses, patenting groups of antibodies will get much harder, especially for research teams that aren’t bankrolled by pharmaceutical companies and represented by high-dollar legal teams.
But it’s not clear yet that the high court will even take up the case, Amgen Inc. v. Sanofi et al. Some experts predict the precedent-loving court will send it back to the lower court. Others see it as a potentially important test case.
(Update 1/8/2019: The Supreme Court Amgen’s petition for review of the appeals court’s decision. The case returned to the District Court of Delaware. For subsequent updates, click here.)
Fifteen years ago, PCSK9 was a barely known gene defined by homology to a family of proteases and by an interesting phenotype in a few humans with mutations. It developed with unusual speed into a drug target and a legal battleground that may set important precedent for the biotech industry.
A new genetic link to cholesterol
The story starts with the discovery that some patients with familial hypercholesterolemia, or very high levels of low-density lipoprotein cholesterol, also known as LDL, carried mutations in a newly identified gene called PCSK9. A group of French scientists led by Catherine Boileau published that finding in the journal Nature Genetics in 2003.
Scientists all over the world took notice. Within a year and a half, three groups had shown in mice that overexpression of PCSK9 in the liver raised blood levels of cholesterol because of a reduction in LDL receptors in the liver.
 Tom Lagace
Tom Lagace
Tom Lagace, an American who had just finished his Ph.D. at Dalhousie University in Canada, joined one of those research groups at the University of Texas Southwestern Medical Center in 2004. He had hoped to focus on transcription factors called SREBPs, but his mentors had a different project in mind for him.
Lagace recalls a senior scientist sitting him down and saying, “‘No, you’re not going to work on that project. You’re going to work on this, PCS, PSC’ — he had to go and get a paper, then he said, ‘Oh, yeah, PCSK9.’”
 Michael Brown (left) and Joseph Goldstein
Michael Brown (left) and Joseph Goldstein
UTSW’s department of molecular genetics had a storied history in cholesterol research. It was home to the lab of Nobel laureates , who had discovered the LDL receptor.
The receptor, expressed on the surface of liver cells, ferries low-density lipoproteins rich in cholesterol from the blood into liver cells through endocytosis. The LDL receptor then is recycled to the cell surface, while the lipoproteins are metabolized. Mutations in the receptor that disrupt this metabolic pathway are linked to high blood cholesterol. Brown and Goldstein’s description of this cycle had paved the way for the development of a class of drugs called statins, and they shared a Nobel Prize for the work in 1985.
Brown and Goldstein continued to research mechanisms of cholesterol homeostasis in a growing department at UTSW, which Goldstein chaired, staffed by many of their former trainees.
 Jay Horton
Jay Horton
Lagace went to work for one of Brown and Goldstein’s former postdocs, Jay Horton, by then a UTSW professor. Horton had come across PCSK9 independently in published in 2003, finding that its RNA level responded to SREBP.
After that study and Boileau’s were published, the Horton lab began to investigate mice that overexpressed PCSK9, which had higher cholesterol.
Horton shared his study results with UTSW colleagues Helen Hobbs, a physician-scientist who had also trained in the Brown and Goldstein lab, and Jonathan Cohen. Hobbs and Cohen had just established the Dallas Heart Study, using population genetics to search out rare genetic variants with significant effects on cardiovascular disease.
 Jonathan Cohen and Helen Hobbs
Jonathan Cohen and Helen Hobbs
After hearing about the effects of PCSK9 overexpression in mice, Hobbs said, she and Cohen decided to look for mutations in PCSK9 that would kill its function. They reasoned that such mutations should appear in people with more LDL receptor activity and lower blood cholesterol.
“We went to our (heart study) population and sequenced the coding regions of PCSK9 in the individuals with the lowest LDL levels and hit pay dirt right away when we found a nonsense mutation,” Hobbs told in 2015.
The study subjects whose PCSK9 genes included a nonsense mutation had LDL levels much lower than those without the mutation. The team also discovered a missense mutation that changed a single amino acid in PCSK9. They tested the effects of these sequence variations in a different group and found that people with PCSK9 mutations had a .
Meanwhile, the Horton lab continued to investigate the molecular mechanisms that led to PCSK9’s effect.
“I was quite lucky that a former postdoc was just leaving” the lab, Lagace said, “so I came at the right time.” His predecessor left him a collection of plasmid constructs and a big question to work on: How did PCSK9 reduce cell-surface levels of the LDL receptor?
PCSK9 is a member of a family of proteases, enzymes that break down other proteins. It stood to reason, and some mechanistic work from other labs suggested, that its effect on cholesterol came through breaking down another protein — but which? What was PCSK9’s substrate?
Lagace set about finding out.
‘Bingo. I had it.’
He did not have much luck.
“My project was to develop a protease assay and find out what PCSK9 (was) cutting,” Lagace said. “And I was getting nowhere. Now, looking back, I was getting nowhere because it’s not active. But at the time … I was really struggling.”
Frustrated and increasingly worried that he had nothing positive to present at the lab’s weekly meeting, Lagace decided to try isolating the protein from mammalian cells instead of the bacterial or insect cells he had been using. Post-translational protein modifications are known to differ among species; perhaps, he thought, some crucial tweak to PCSK9 was missing in the protein purified from bacteria.
After setting up a system in the lab to express and purify the protein from human cells, he said, “I was really struck that the PCSK9 in the medium was much greater than the PCSK9 in the cells.” That led him to hypothesize that the protein is mostly secreted — and that the previously published work suggesting PCSK9 operated inside of cells was somehow incorrect.
To test the idea, Lagace asked a nurse who worked nearby to draw some of his blood. He checked it for PCSK9; the protein was definitely in his plasma. Then he asked a colleague who processed and fractionated patient samples to run his plasma through a size-exclusion column, which separates macromolecules in a solution according to their mass.
“All I was going to do was see how big PCSK9 was (in case it oligomerized), but he also gave me the lipoprotein profiles. I was really struck that a lot of the PCSK9 was in a perfect overlap with low-density lipoprotein in the blood,” Lagace said. A quick experiment using isolated proteins and lipoproteins confirmed that PCSK9 bound to low-density lipoprotein particles.
“Nobody knows this, but the first thing I ever found wasn’t LDL receptor binding; it was LDL binding.”
Thinking that PCSK9 might piggyback on LDL particles and attack LDL receptors once it was internalized, he set up an experiment controlled with PCSK9 alone in LDL receptor-positive and -negative cells. To his surprise, even without LDL, PCSK9 could make it into the cells — but only if the receptor was present.
“So that was it. Bingo. I had it,” Lagace said. “(PCSK9) binds to the LDL receptor.”
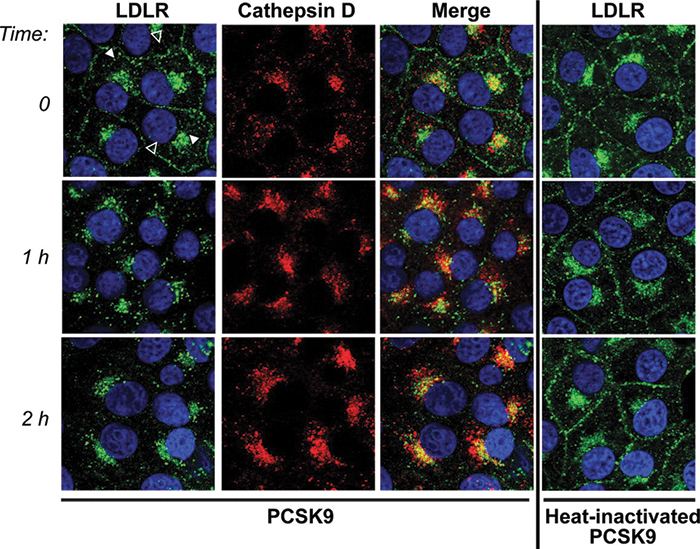 An imaging experiment from a Horton lab publication shows the effect of PCSK9 on LDL receptor internalization. The receptors (green) move into lysosomes (marked with cathepsin D, red) after PCSK9 is added. If PCSK9 is denatured (right column), the internalization does not occur. Zhang et al./ Journal of Biological Chemistry
An imaging experiment from a Horton lab publication shows the effect of PCSK9 on LDL receptor internalization. The receptors (green) move into lysosomes (marked with cathepsin D, red) after PCSK9 is added. If PCSK9 is denatured (right column), the internalization does not occur. Zhang et al./ Journal of Biological Chemistry
His project took off. The next steps were clear, and the Horton lab, with the Hobbs and Cohen lab, was on the way to publishing a in the Journal of Biological Chemistry defining the mechanism by which PCSK9 affects LDL receptors.
In short, circulating PCSK9 binds to the LDL receptor’s extracellular domain. When the LDL receptor is internalized by endocytosis, PCSK9 directs it to the lysosome for degradation. If circulating PCSK9 is blocked, the number of LDL receptors on the surface of liver cells goes up — providing a way to remove more LDL cholesterol from the blood.
“Because PCSK9 is secreted in the blood and works extracellularly,” , “an opportunity to create antibodies was created. These antibodies can function in the blood and block the interaction of PCSK9 with the LDL receptor, essentially resulting in an inhibitor of the PCSK9 protein.”
Meanwhile, at Amgen
The potential value of a PCSK9 inhibitor was clear both to Horton and to scientists at pharmaceutical companies. When the 2003 Nature Genetics paper came out, Simon Jackson was working at a biotechnology startup called Tularik. He read the paper, chatted with his supervisor about it and put it away. It wasn’t the right time, he said in court in 2016.
In 2004, Amgen bought Tularik. Jackson stayed on as a scientist. PCSK9 came back to his attention in 2005, when Goldstein visited Amgen and gave a lecture on findings from the Horton lab.
Horton’s group had found not only that mice without PCSK9 had lower cholesterol but also that giving them blood from a mouse that did carry the gene raised cholesterol again, suggesting that PCSK9 operates in the bloodstream and not within liver cells. Hobbs and Cohen’s discovery that humans with nonfunctional PCSK9 have lower cholesterol was not yet published, but Jackson saw evidence in Goldstein’s talk that that reducing PCSK9 in the blood could be beneficial to patients.
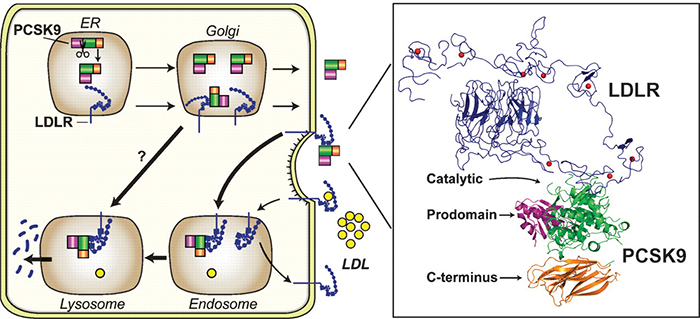 A figure from a review article by Horton, Cohen and Hobbs shows how PCSK9 (green, purple and orange) promotes internalization and destruction of the LDL receptor (blue) in liver cells. Horton et al./Journal of lipid research
A figure from a review article by Horton, Cohen and Hobbs shows how PCSK9 (green, purple and orange) promotes internalization and destruction of the LDL receptor (blue) in liver cells. Horton et al./Journal of lipid research
“So I presented to the senior team at Amgen,” Jackson testified in 2016. He earned their approval to begin research on ways to block PCSK9 activity, focusing on its hypothetical protease activity.
At around the same time Lagace was making his discoveries, Jackson and his team at Amgen were arriving at the same conclusion: PCSK9 bound directly to the LDL receptor, reducing its presence on the surface of liver cells.
Was the discovery completely independent?
At trial, Sanofi’s lawyers pointed out that Horton, like Goldstein, had visited Amgen while this research was ongoing. Amgen’s witnesses insisted that the scientific discussions concerned only published work. (Cohen and Goldstein both declined requests to be interviewed for this story; Horton and Hobbs did not respond.)
In any case, once it was clear that PCSK9 functioned by binding to the LDL receptor, and not as an enzyme, the next step was to come up with a protein product that could block that binding. By doing that, researchers could block LDL receptor internalization and degradation, leaving the protein able to suction more LDL out of the bloodstream. That would lower patients’ risk of heart attack.
To find proteins that block PCSK9 from binding to the LDL receptor, Jackson called on Amgen’s antibody team, led by Chadwick King.

“What we wanted to do was design an antibody that bound to the LDL receptor binding site (on PCSK9) and blocked that interaction,” King said at trial. “What we didn’t know was exactly where the site was on the molecule.”
With a clever assay design, however, the team didn’t need to know the exact binding location. They just needed to find out which of about 3,000 antibodies they had raised against PCSK9 in mice could block the protein from also binding the LDL receptor.
“What we were asking was, if we had PCSK9 in solution and we allowed that (experimental) antibody to bind, could that complex then bind to the LDLR on the plate?” King explained at trial.
The team found 85 antibodies that block PCSK9–LDL receptor binding. They continued to characterize that subset, looking for antibodies that would make promising drug targets.
The race to the patent office
The Amgen team was by no means the only industry research group working on PCSK9; the Amgen process is unusually public because they later disclosed it all in court. After Hobbs and Cohen published their finding that loss of PCSK9 lowered LDL cholesterol, it was clear the protein would become an important drug target. Hobbs told the , “There were a number of drug companies that picked it up.”
Papers on protein crystal structures and patent applications indicate that teams at Novartis, Pfizer and Regeneron also were hard at work trying to block PCSK9. Adding urgency to their studies, everyone knew that statins, the blockbuster cholesterol-lowering drugs that included Lipitor, soon would come to the end of their patent protection and become much less profitable.
 Charles Craik
Charles Craik
Charles Craik, a professor of pharmaceutical chemistry at the University of California, San Francisco, who doesn’t work directly on PCSK9 but has followed its emergence as a drug target, said that with statins going off patent, “the whole market was wide open for something new. … There was a big race, and there was a lot of money involved.”
Amgen won the race to the patent office. It took Jackson’s team about a year and a half, from June 2006 to January 2008, to collect all the data they used in a patent filing. Beginning late in 2007, Amgen filed 30 patents for antibodies to PCSK9 with the U.S. Patent and Trademark Office, and a total of .
 Ari Zytcer
Ari Zytcer
According to Ari Zytcer, an intellectual property lawyer and trained pharmacist based in Ohio who prepares patent applications, that flurry of patents is a typical strategy.
“Any time there is a drug that is this important, (lawyers) build a patent estate around it,” said Zytcer, who does not represent Amgen. “You don’t want to just cover the invention in a single way. You claim it in as many ways as you possibly can.”
The most important of Amgen’s patents, filed in January 2008, described the screen that King and colleagues had performed to winnow the 3,000 antibodies to a few leading drug candidates. The patent laid claim to a class of antibodies that block PCSK9’s interaction with the LDL receptor. The patent filing also included crystal structures of two of those candidates in complex with PCSK9 and amino acid sequences of about two dozen more.

The two runners-up, Regeneron and Pfizer, filed applications before the end of 2009 to patent antibodies to PCSK9 to treat high cholesterol. Regeneron, the smallest of the three companies, licensed its product to pharmaceutical giant Sanofi for clinical development and commercialization.
In the race to launch a PCSK9 inhibitor, three of the year’s top 15 drug companies were now in the running.
By the fall of 2014, both Amgen and Sanofi had demonstrated that their PCSK9 inhibitor antibodies were safe and effective treatments for patients with elevated LDL cholesterol, and both companies had submitted data from large efficacy trials to the Food and Drug Administration. They expected final FDA decisions around the same time on whether they could sell the molecules as drugs. Pfizer was a few years behind them, and later scuttled its drug candidate.
That October, the patent office finally approved Amgen’s 2008 application on its PCSK9 inhibitor antibody screen. Just days later, Amgen sued Sanofi for patent infringement.
Why did Amgen sue?
The patents Amgen held, the company’s lawyers said, described a whole class of antibodies — any antibody that would bind to the LDL receptor-binding region of PCSK9, often referred to in court as the protein’s “sweet spot.” Sanofi was preparing to market an antibody that bound exactly there, meaning it fell into the protected class. Amgen’s lawyers insisted that Sanofi stop.
Sanofi hired the best possible lawyer for its particular case. Dianne Elderkin, a patent litigator in Philadelphia, had represented Janssen Biotech in an important settled earlier that year. Abbvie, which owned the patent on a class of antibodies against a cytokine, had sued Janssen for developing an antibody that binds to the same target. Elderkin led the legal team defending Janssen from charges of patent infringement. Her team’s case convinced the jury to find Abbvie’s patent invalid.
Sanofi faced a problem similar to Janssen’s: It was being sued for infringing on a patent that claimed a class of antibodies. Working as lead counsel with a team of lawyers and paralegals from three law firms, Elderkin became the face of Sanofi’s argument that Amgen’s patent was invalid.
Squaring off against Elderkin, Amgen’s lead counselors were Sarah Columbia, a Boston litigator whose prior experience tended more to devices than large molecules, and William Gaede, a colleague at Columbia’s firm who was experienced in biotech. The two headed up a team of lawyers and paralegals from two law firms.
Lawyers on both sides began to review each company’s laboratory records, recruit professors to serve as expert witnesses, build their arguments and dispute which evidence was admissible in court.
As the case lumbered toward a trial, the FDA approved both drugs for the treatment of high LDL cholesterol in 2015. The two drugs launched commercially, Sanofi’s as Praluent and Amgen’s as Repatha, about a month apart that summer. Less than six months later, the patent-infringement suit went to trial in Delaware as Amgen Inc. v. Sanofi et al.
Pretrial negotiations were contentious, and neither side gave any ground. The presiding judge was , a 24-year veteran of Delaware’s District Court, one of the country’s busiest intellectual property arenas. During a bench hearing early in the proceedings, Robinson observed to the opposing legal teams, “You all are going to appeal no matter what happens in this case, whether I stop it now, whether we go to a verdict, whatever I do with the verdict, whatever the jury does with the verdict.”
The arguments
The legal issues were dense, but Amgen’s core argument was simple. According to trial records, its lawyers argued that Sanofi and Regeneron had infringed on two of Amgen’s patents, which protect any antibody that targets a specific LDL receptor-binding region of PCSK9. (Amgen’s media relations office did not respond to a request for comment).
Sanofi’s defense was not that its product didn’t infringe on Amgen’s patents. Instead, lawyers argued that the claims were inappropriately broad. In a statement to 91—«…´¥´√Ω Today, a Sanofi spokesperson wrote, “Amgen’s patent claims essentially cover any anti-PCSK9 antibody that would work. Sanofi and Regeneron contend that these patent claims are invalid.”
Everyone agreed that if the patent was valid as written, Sanofi had infringed upon it, so the issue before the jury was whether the patent was valid. In other words, did Amgen own every monoclonal antibody that binds the LDL receptor-binding region in PCSK9? To answer that question, the jury first needed to decide whether Amgen had described adequately a class of antibodies and how to make them.
The law states what can’t be patented: You can’t patent something that’s obvious, something that occurs naturally or — in most cases — an idea that hasn’t been made real.
A patent needs to include a description of the product and a reasonable amount of information about how to make it, called enablement. These rules are a little relaxed for antibodies.
Also important in this case is that an inventor can patent a class of products — provided the patent application discloses enough examples in the class’ written description that a colleague could see how they’re related and imagine the rest of the class.
Being able to patent a class is useful in biotechnology. Otherwise, a competitor could tweak just enough of an antibody’s sequence to fall outside of a patent’s claims, profiting off an inventor’s work. But it also raises a problem, because imagining all of the possible antibodies that could bind to a given epitope — all of the members of a class — is all but impossible.
 Many types of antibodies, including most of the monoclonal antibodies used to treat diseases, share two polypeptides the heavy and light chains, arranged in a Y-shaped structure. The specificity of antibody binding comes from sequence variation that leads to structural variation in the variable region, here indicated in purple. Wikimedia/Digital Shutter Monkey
Many types of antibodies, including most of the monoclonal antibodies used to treat diseases, share two polypeptides the heavy and light chains, arranged in a Y-shaped structure. The specificity of antibody binding comes from sequence variation that leads to structural variation in the variable region, here indicated in purple. Wikimedia/Digital Shutter Monkey
In an influential antibody case decided in 2011, the U.S. Court of Appeals for the Federal Circuit described an epitope not as a lock with a single key, but a lock and “a ring with a million keys on it.” And legal precedent, including the case Elderkin had won for Janssen, held that to own a class of antibodies, the inventor needs to describe a broad cross-section of the class, representing the diversity of those keys.
Sanofi’s lawyers contended that Amgen had not provided enough information about how its family of antibodies was made or description of the antibodies themselves to qualify as a thorough description of the class. The team argued that the antibodies disclosed in the patent bound only to the edges of the PCSK9–LDL receptor binding site and, more importantly, that there was no way to envision what other antibodies would fit into the class.
Besides written description and enablement, one more important legal concept was in play. At the time Amgen had filed its patent, the part of PCSK9 that bound to the LDL receptor was not widely known. That meant Amgen’s claims were also subject to a patent office assessment called the newly characterized antigen test, which protects inventors who find antibodies binding to a new and important antigen, even if the rest of the invention is routine.
For five days, the opposing legal teams and their expert witnesses argued over whether Amgen’s patents fulfilled the written description and enablement requirements. Near the end of the trial, during an argument at the bench about how to phrase the jury’s instructions for deliberation, Judge Robinson said, “This is the most complicated trial I’ve ever had.”
One win each
Amgen convinced the jury that its patent claims were valid. The U.S. District Court for the District of Delaware ruled that Amgen’s patent was indeed valid, and Robinson issued an injunction that would stop Sanofi from selling its PCSK9 inhibitor, Praluent. The website Law.com named Columbia and Gaede for the victory.
Just as Robinson had predicted, Sanofi immediately appealed. The U.S. Court of Appeals for the Federal Circuit took up the case and suspended the injunction before it went into effect. During the appeal, Sanofi parted ways with Elderkin’s firm. Amgen added a few lawyers but kept its trial team otherwise intact.

In October 2017, the appeals court overturned parts of the lower court’s ruling. It concluded that the lower court had been wrong to exclude some information that Sanofi had hoped to use to convince the jury that Amgen’s written description and enablement were insufficient. That information, from Regeneron’s patent application, was intended to illustrate the gap between what Amgen disclosed and the antibodies that subsequently were discovered to bind to PCSK9. It is key to a thorny question: What if a team of inventors discloses everything they have discovered, but they haven’t yet discovered everything?
The appeals court also noted that experts in the trial had not agreed on whether knowing an antigen’s characteristics enabled scientists to make antibodies to it. In its decision, the court wrote that “it has been, at the least, hotly disputed that knowledge of the chemical structure of an antigen gives the required kind of structure-identifying information about the corresponding antibodies.”
In light of that ongoing dispute, the appeals court ruled that describing an antigen could not suffice as written description of the antibody that binds it.
 Irena Royzman
Irena Royzman
That ruling will guide the judge who hears the case if it returns to the lower court in Delaware. Judge Robinson has since retired; at the retrial, scheduled for February, the new judge cannot tell the jury that written description of an antigen suffices to describe the antibody it binds. The jury will consider the same question: Does Amgen’s disclosure of 384 PCSK9 inhibitors that block the protein’s interaction with the LDL receptor describe a class of antibodies well enough to claim it?
Irena Royzman is a New York patent lawyer who earned a Ph.D. in biology before law school and now litigates biotechnology cases. “The question before the court at the district court level will be whether what Amgen discloses, under the law as now clarified by the federal court, is sufficient,” Royzman said. “The question is: How broad is the claim? How substantial is the disclosure in light of the breadth of the claim, and do they really have representative species?”
But whether the retrial will go forward as scheduled is uncertain. In July, Amgen appealed to the U.S. Supreme Court to review the federal appeals court’s decision.
The highest court
“In Amgen’s petition to the Supreme Court, they’re essentially raising a new issue,” Royzman said.
“They’re saying that the federal circuit is completely confused.”
Intellectual property lawyer Ari Zytcer elaborated on Amgen’s position, explaining that the company argues that the appeals court misinterpreted the patent law. Amgen’s lawyers, he said, contend “that the current interpretation by the federal circuit court of a separate written description and enablement requirement is actually improper … (and) in fact it’s a single requirement.”
Other pharmaceutical companies, notably Bristol–Myers Squibb, weighed in with an amici curiae (“friends of the court”) brief, arguing the Supreme Court should hear the case. The companies argued that the appeals court’s ruling impedes innovation, because once an inventor has one antibody to an epitope, it’s trivially easy to make what they called follow-on antibodies. Moreover, the brief said, compiling a “representative number of examples” could be quite subjective — just how many examples is that? — and might involve a lot of rote work.
Royzman doubts that the Supreme Court will take up the case. “(Amgen’s lawyers) are arguing that there is no such thing as the written description requirement: that the Federal Circuit just came up with it and it’s not grounded in a statute. I don’t think that’s correct.”
On Nov. 19, Sanofi filed a response to Amgen’s Supreme Court petition, calling the case “an exceptionally poor candidate” on the grounds that Amgen had never questioned the interpretation of patent law in the lower courts. Moreover, Sanofi argued, even if the Supreme Court did review how the appeals court is applying the law, it would not settle the central question of whether Amgen’s patent is valid. Now it’s up to the Supreme Court to decide whether to intervene in the case.
Whether the case appears clear-cut could depend partly on whether a lawyer writes patents or litigates them, Zytcer noted. “From my perspective, I’d like the Supreme Court to chime in.”
He sees uncertainty in how patent law is interpreted among legal practitioners and patent examiners that could be settled by a Supreme Court decision. “I think from a litigator’s perspective, though, this is plain and simple.” There is plenty of precedent upholding separate enablement and written description requirements for patent applications, he said.
“Amgen really did describe more than most applicants would ever put into a patent application,” Zytcer said. “So the fact that their patent could be held invalid on the basis of lack of written description … that would have a chilling effect on the biotech industry.”
The case has major implications for a type of patent dispute that comes up again and again — and is poised to become even more common as antibody drugs proliferate. Not long after the Delaware court’s initial decision in Amgen v. Sanofi, Merck and Bristol–Myers Squibb settled over antibodies against the cancer antigen PD-1. Merck paid Squibb $625 million and will pay royalties on its drug Keytruda for the next eight years.
At least two other cases await litigation to determine whether competitors’ drugs infringe on patents covering classes of antibodies defined by antigens. Bristol–Myers Squibb, one of the friends of the court advocating for the Supreme Court to hear Amgen’s case, recently hired Elderkin and her team to sue AstraZeneca over PD-L1 inhibitors, which target PD-1’s ligand and are also used to treat cancer. In July, Genentech against Eli Lilly and Co. over Lilly’s interleukin-17 antibody just after Genentech’s patent for a class of humanized monoclonal antibodies to the interleukin was granted.
The increasingly crowded monoclonal market
As antibody drugs proliferate, there is more and more opportunity for companies’ patent claims to overlap. The number of FDA-approved antibody drugs has surged in recent years, and many are in legal dispute. Challenges concerning approved drugs that target the EGF receptor, cytokine IL-12 and cancer-related receptor Her2 have shaped the patent landscape, and many infringement cases concerning drugs to other targets are ongoing.
In 2017, the U.S. Patent and Trademark Office granted 964 patents with titles that contained the words “antibody” or “antibodies.” Those titles sometimes described a broadly applicable method or specified a drug-antibody conjugate, but the majority simply described an antibody or group of antibodies and left the details for the claims.
Only about 2 percent of U.S. patents granted in 2017 were assigned to the world’s largest pharmaceutical companies by revenue — a group to which both Amgen and Sanofi belong. About a quarter were assigned to universities or academic–commercial partnerships, and more than half went to smaller biotech and pharmaceutical companies, and in some cases individual inventors, from around the world. Academic and small commercial inventors may lack the resources to protect their intellectual property with the vigor that Amgen and Sanofi have shown in litigation.
A Supreme Court decision concerning Amgen v. Sanofi could reach beyond these cases, according to Anna Lukacher and Richard Kurz, who work on pharmaceutical patents in New York. The Supreme Court selects cases “with the intent to clarify the law,” Kurz said. “And they tend to rule in a way that is interpreted very broadly. Any time the Supreme Court takes on a case, it usually has an impact on a wide variety of cases going forward.”
According Lukacher and Kurz, the retrial at the lower court in Delaware and consideration at the Supreme Court level will proceed in parallel unless the judge in Delaware chooses to stay the trial — which Kurz said would be likely if lawyers for Amgen or Sanofi asked for a pause.
“I would say it’s unlikely that the Supreme Court would get its done and actually take up the case in time for the district court judge (in Delaware) to have any reason to stay the trial,” he said. “That’s reading the tea leaves a little bit, but that’s faster than the Supreme Court usually moves.”
The future of antibody patents
The U.S. Patent and Trademark Office has taken what steps it can to prevent a repeat of this battle. In a memo issued after the appeals court made its decision, the patent office instructed patent examiners that “adequate written description of a newly characterized antigen alone should not be considered adequate written description of an antibody to that antigen,” officially putting the appeals court’s ruling into practice at the patent office.
However, lawyers say there is still confusion among patent examiners about what suffices as description.
Zytcer said of a recent patent office rejection, “Examiners and the legal folks at the patent office can’t give a straight answer because (the law) is just open to too much interpretation.”
Amgen’s petition to the Supreme Court argued that the appeals court’s ruling has “left innovators no way of predicting what disclosures will be sufficient” to secure patent protection for their inventions.
“I think that’s somewhat exaggerated,” Royzman said about Amgen’s assertion. “For example, if you write a claim that’s specific to your antibody, you can obviously get that claim. If you write a claim that says ‘my antibody with sequence variation of 80 or 90 percent,’ people are getting claims like that.”
On the other hand, she said, “If you’re going to say ‘any antibody that binds to receptor X or protein Y, those are all mine,’ then those claims are highly problematic.”
Charles Craik, the UCSF professor, holds . “I find it very disappointing that that’s the direction antibody patents are going,” he said. “It’s slicing the salami so thin that you can’t get a meal off of it.”
For example, he said, his colleague , who is now vice chancellor for business development at UCSF, the mouse antibody. “That’s a spectacular , one of the most significant in biotechnology,” Craik said. “If you’re slicing things as thinly as the outcome of Amgen v. Sanofi, you couldn’t get that patent.”
Craik once wrote an application for a class of antibodies that was denied on the grounds that it claimed more than he possessed when he submitted the application. (He had hoped to claim antibodies that could bind to any protease and prevent conformational shifting from an active to an inactive form, but he secured a patent for antibodies against the active form of just one protease.) He worries that narrowing the scope of the average patent to protect specific epitope-binding sequences will harm the commercial possibilities of monoclonal antibodies.
According to Zytcer, if the appeals court’s ruling holds, it may dampen universities’ enthusiasm for patenting antibodies, especially for technology transfer offices with limited resources.
Lawyers writing patent applications that describe antibodies, he said, are “going to try to add a little more disclosure — but in reality, you’re not going to get anything else from the clients. They have what they have at the time of development.”
All the same, Royzman said, there's a strong argument to be made in Sanofi’s favor — one she said the appeals court’s ruling takes into account. “It is an important decision, because look: A lot of innovators develop their own antibodies, and it’s a lot of work,” Royzman said. “And those antibodies with their specific sequences are beneficial to patients and beneficial to us as a society — and it’s problematic if somebody owns a target. That’s at core what this case is about.”
(Update 4/30/2019: In February 2019, a jury in the District Court of Delaware ruled that two of the five claims in Amgen’s two patents under dispute were invalid and three were not. However, the Sanofi’s argument that Amgen provided too little information about how to make the antibody.
Since everyone agreed that if the patents were valid, Sanofi was infringing on them, Amgen has asked the court for a permanent injunction against sales of Sanofi’s medicine, Praluent. Sanofi has requested a new trial on procedural grounds related to what evidence was permitted and how the jury was instructed in the retrial. Both the motion for a new trial and the injunction hearings are still under consideration as of April 30.)

What’s in a patent?
From a lawyer’s point of view, a patent is essentially a trade. An inventor tells the government what they have invented and how it’s made; the federal government protects the inventor’s exclusive rights to the invention for a set amount of time, usually 20 years.
What goes into the application is governed by a law that reads, in part, “The specification shall contain a written description of the invention, and of the manner and process of making and using it, in such full, clear, concise and exact terms as to enable any person skilled in the art to which it pertains, or with which it is most nearly connected, to make and use the same.”
In most fields, that means a structural definition of the product, perhaps with some diagrams.
As anyone who has taken a biochemistry course knows, protein sequence determines structure, and protein structure determines function. So for protein inventions, applications that include the protein’s amino acid sequence or its function have been accepted as reasonable proxies to structure. This is especially true for monoclonal antibodies, which are invented specifically to bind to other proteins.
Until recently, the U.S. Patent and Trademark Office would consider granting a patent application that described in detail the function of an antibody — by describing the epitope it bound. This was known as the “newly characterized antigen test.” The Amgen v. Sanofi patent-infringement case, however, has forced the patent office to stop using that test.

The origin of antibodies
 Nobel laureate César Milstein invented an efficient method for generating monoclonal antibodies, which he never patented. UK Medical Research Council
Nobel laureate César Milstein invented an efficient method for generating monoclonal antibodies, which he never patented. UK Medical Research Council
Before scientists developed methods to produce monoclonal antibodies, researchers who needed antibody activity relied on antisera, heterogeneous mixtures of antibodies isolated from immunized animals. In 1975, Argentinian biologist developed a method to make a single antibody species, or monoclonal antibody, by fusing a single antibody-producing cell from an immunized animal with an immortalized cancer cell. Milstein shared the 1984 Nobel Prize in Medicine for the finding.
Although the invention, called a hybridoma, spawned drug industry profits now estimated at $90 billion a year, Milstein never applied to patent it.
That decision was famous and controversial. Though Milstein did go into business with a local antiserum producer to sell monoclonals, the outfit couldn’t compete with larger biotechnology companies. He developed the method while working at the Laboratory of Molecular Biology in Cambridge, England. British Prime Minister Margaret Thatcher criticized what she called Milstein’s failure to patent the invention. Milstein, who had disclosed the hybridoma technology to his funding agency before publishing it in the journal Nature, blamed the bureaucracy for not pursuing a patent.
Charles Craik, a professor of pharmacology at the University of California, San Francisco, holds 16 patents. “It’s amazing that C√©sar Milstein didn’t try to patent monoclonal antibodies, hybridomas,” Craik said. “He just published that material, and the world benefited enormously.”

Overheard in the courtroom
When Amgen Inc. v. Sanofi et al. went to trial in the District Court of Delaware, expert witnesses — many of them members of the 91—«…´¥´√Ω — gathered at the courthouse in Dover to explain the scientific dispute to the jury and give their opinions on whether Amgen held a valid patent. A selection of their statements, in which they tried to bridge the legal and biochemical worlds, is offered below.
 Sarah Columbia, attorney for Amgen:
Sarah Columbia, attorney for Amgen:
“Would you agree with me or not that these authors … concluded that the molecular mechanism of PCSK9 action on the LDL receptor and the relationship of their observations remained to be determined?”
 Jeffrey Ravetch, professor at the Rockefeller University:
Jeffrey Ravetch, professor at the Rockefeller University:
“They state that. And you have to unpack that a bit because we write in jargon to each other. What they are saying is, we have identified LDLR as being an important downstream molecule for PCSK9. The precise molecular details we may need to determine. They put the second important piece in place but didn’t know exactly the conclusions. That’s the way science works. We build on top of prior observations.”
 “I’m not going to make any particular orientation here, but let’s just say it binds like this. Okay? Then you can see if it does, you won’t be able to get the LDL receptor in. That’s what we mean by blocking. … You can see my assistant here is unable to get the LDL receptor to bind.”
“I’m not going to make any particular orientation here, but let’s just say it binds like this. Okay? Then you can see if it does, you won’t be able to get the LDL receptor in. That’s what we mean by blocking. … You can see my assistant here is unable to get the LDL receptor to bind.”
— Gregory Petsko, professor of neurology and neuroscience at Weill Cornell Medical College, while showing 3D printed molecule models based on the crystal structures in Amgen’s patent
 “I expect that there are antibodies — many, many antibodies — that will meet Amgen’s claims that have nevertheless very diverse and different three-dimensional structures and primary amino acid sequences. I couldn’t begin to tell you, by looking at that structure, what those particular amino acid sequences are or what those three-dimensional structures are other than the two examples that Amgen has provided in these patents.”
“I expect that there are antibodies — many, many antibodies — that will meet Amgen’s claims that have nevertheless very diverse and different three-dimensional structures and primary amino acid sequences. I couldn’t begin to tell you, by looking at that structure, what those particular amino acid sequences are or what those three-dimensional structures are other than the two examples that Amgen has provided in these patents.”
— Michael Eck, professor of biological chemistry and molecular pharmacology, Harvard Medical School
 “Granted, it is hard to get a straight answer out of experts on a general basis, but I believe he said there are different ways of doing it and he was just relying on what the inventors in this case did.”
“Granted, it is hard to get a straight answer out of experts on a general basis, but I believe he said there are different ways of doing it and he was just relying on what the inventors in this case did.”
— Delaware District Court Judge Sue Robinson, ruling on whether an expert witness Donald Siegel had contradicted a prior deposition
 “All this talk about ‘We found a sweet spot’ adds absolutely nothing to the knowledge of people of skill in the art going to make antibodies. It’s just like, ‘go make more, and then if you find one that binds here, great for you; that falls within our claims.’ ”
“All this talk about ‘We found a sweet spot’ adds absolutely nothing to the knowledge of people of skill in the art going to make antibodies. It’s just like, ‘go make more, and then if you find one that binds here, great for you; that falls within our claims.’ ”
— Dianne Elderkin, attorney for Sanofi, during a bench conference, on whether Amgen’s disclosures had enabled other scientists to generate all antibodies in the class without undue experimentation

Enjoy reading 91—«…´¥´√Ω Today?
Become a member to receive the print edition four times a year and the digital edition weekly.
Learn moreGet the latest from 91—«…´¥´√Ω Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Predicting fatty liver disease from a tiny blood sample
Obesity and being overweight aren't the only factors that contribute to liver disease. New tests can help identify who is at risk or already has the disease, even in people who are lean or have a normal weight.

An ancient animal helps scientists improve modern technology
The same molecules that help tardigrades survive extreme weather can improve cryo-EM images of cellular structures and proteins, a team led by University of Wisconsin–Madison researcher Ci Ji Lim reports.

New structure gives insight into mRNA export and cancers
Yi Ren’s lab at Vanderbilt has described the structure of a protein complex that sheds light on the underlying molecular mechanism of mRNA export.

Analyzing triglycerides in Americans of African ancestry
Using the All of Us database, researchers at Vanderbilt sought a genetic reason why some patients, often underrepresented in research, could have varying levels of fat in the bloodstream.

Of yeasts and men: One-hour proteomes, 10 years apart
To profile the human genome within an hour, the researchers used a new mass spectrometer and packed their liquid chromatography columns with very high pressure.

Cells have more mini ‘organs’ than researchers thought
Membraneless organelles, also called biomolecular condensates, are changing how scientists think about protein chemistry, various diseases and even the origin of life.

